Beware of powder assemblies for two plants of the same condition and outliers
Note that when I pooler I consider twice the same value for two different plants. Ex: 1067 and 1069 pooler. The result goes to 1067 and 1069 not to 1067&1069)
I also removed the two plants 1039 and 1087 on 14/12/2022 for random draws (two plants out of type).
11.2 Efficicence with bootstrap
11.2.1 Function for bootstrap EUE & sREU
Function for Element Use Efficiency based on random drawing between plant at harvest 1 (control plant) and plant at harvest 2 (control or also treated condition). Each time we combine the same genotype. Idem for specific Root Element Uptake but is an integration during between date and we use root weight.
Code
# warning, concentration in µg/g en root in g. random_drawing_iono_EUE_sREU<-function(rep){set.seed(1) df_compile=data.frame()#do it for each compound!!! #Do not take more than one organ because we are looking at the total quantity of an element df_ionomic_select=df_ionomic %>%filter(compartiment=="leaf") %>%drop_na(sum_by_component) %>%filter(compound!="Sb121") %>%filter(num_compartiment==3) %>%drop_na(weight_total) %>%filter(sum_by_component>0) %>%filter(!plant_num%in%c(1039,1087)) list_condition=c("Sto_WW_OT","Sto_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS")for(e inlevels(as.factor(as.character(df_ionomic_select$compound)))){tic()for(cond in list_condition){for (i in1:rep){#for R2 df_r2=df_ionomic_select %>%filter(recolte=="2") %>%filter(compound==e)%>%filter(condition==cond) df_r2_taking=df_r2 %>%filter(plant_num==sample(df_r2$plant_num,1,replace=T)) %>% dplyr::select(weight_total,condition,compound,sum_by_component,plant_num,recolte,date_recolte,weight_root)#now for r1if(str_sub(df_r2_taking$condition, 1, 3)=="Sto"){ df_r1=df_ionomic_select %>%filter(recolte=="1") %>%filter(compound==e) %>%filter(condition=="Sto_WW_OT") df_r1_taking=df_r1 %>%filter(plant_num==sample(df_r1$plant_num,1,replace=T)) %>% dplyr::select(weight_total,condition,compound,sum_by_component,plant_num,recolte,date_recolte,weight_root) }else { df_r1=df_ionomic_select %>%filter(recolte=="1") %>%filter(compound==e) %>%filter(condition=="Wen_WW_OT") df_r1_taking=df_r1 %>%filter(plant_num==sample(df_r1$plant_num,1,replace=T)) %>% dplyr::select(weight_total,condition,compound,sum_by_component,plant_num,recolte,date_recolte,weight_root) }colnames(df_r1_taking)=c("weight_total_r1" ,"condition_r1" ,"compound_r1" ,"sum_by_component_r1" ,"plant_num_r1","recolte_r1","date_recolte_r1","weight_root_r1")colnames(df_r2_taking)=c("weight_total_r2" ,"condition_r2" ,"compound_r2" ,"sum_by_component_r2" ,"plant_num_r2","recolte_r2","date_recolte_r2","weight_root_r2") df_bind=cbind(df_r2_taking,df_r1_taking) df_bind$rep=i####################### Element Uptake Eficiency ########################## df_bind$EUE= (df_bind$weight_total_r2-df_bind$weight_total_r1) / (df_bind$sum_by_component_r2-df_bind$sum_by_component_r1)#juste to verify df_bind$EUE_numerator=(df_bind$weight_total_r2-df_bind$weight_total_r1) df_bind$EUE_denominator=(df_bind$sum_by_component_r2-df_bind$sum_by_component_r1)####################### Specific Root Element Uptake ########################## df_bind$sEUpE=(df_bind$sum_by_component_r2-df_bind$sum_by_component_r1)/ (( (as.numeric(as.Date(df_bind$date_recolte_r2)-as.Date(df_bind$date_recolte_r1))) * (df_bind$weight_root_r2+df_bind$weight_root_r1)/2 )) df_compile=rbind(df_compile,df_bind) }print(paste(sep="_",e,cond))}toc() } df_compile$compile_rep=paste(sep="_",df_compile$rep,df_compile$condition_r2)#df_compile =df_compile %>% filter(EUE>0)#write resultwrite.csv(df_compile,here::here(paste(sep="","data/iono/output/random_drawing_EUE_sREU",#as.character(format(Sys.time(), "%Y%m%d_%H%M%S")),"_rep",rep,".csv")))return(df_compile)}
11.2.2 Function execution with 100 repetitions
This function can take a very long time !!!
Code
rep=100df_EUE_sREU=random_drawing_iono_EUE_sREU(rep=rep) #function little bit slow but very usefull
11.2.3 Graphical verification
11.2.3.1 For EUE
I don’t know if it’s right, but sometimes there are negative values.
I removed them to generate the graphs
Code
# Warning, concentration in µg/g en root in g. df_EUE_sREU=read.csv(here::here("data/iono/output/random_drawing_EUE_sREU_rep100.csv"))[-1] #%>% #filter(EUE>0)# Graphics creation and assemblyvector_compound=levels(as.factor(as.character(df_EUE_sREU$compound_r1)))plots <-lapply(1:length(vector_compound), function(i) { df_EUE_sREU_for_plettre<-df_EUE_sREU %>% dplyr::rename(condition=condition_r2) %>%mutate(genotype=ifelse(str_sub(condition, 1,3)=="Sto","Stocata","Wendy")) %>%mutate(water_condition=str_sub(condition,5,6)) %>%mutate(heat_condition=str_sub(condition,8,9)) %>%filter(compound_r1==vector_compound[i]) %>%mutate(condition=factor(condition,levels=c("Wen_WW_OT","Wen_WW_HS","Wen_WS_OT","Wen_WS_HS","Sto_WW_OT","Sto_WW_HS","Sto_WS_OT","Sto_WS_HS")))FigX_stat=stat_analyse(data=df_EUE_sREU_for_plettre %>%mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>%mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>%mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>%drop_na(climat_condition,EUE),column_value ="EUE",category_variables =c("climat_condition"),grp_var ="genotype",show_plot = T,outlier_show = F, label_outlier ="compile_rep",biologist_stats = T,Ylab_i =paste0("EUE for ",vector_compound[i], " (g.µgElement-1)"),control_conditions =c("WW_OT"),strip_normale = F,hex_pallet = climate_pallet)px=FigX_stat[["plot"]] +theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major =element_blank(),panel.grid.minor =element_blank(),legend.position ="bottom",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+labs(color="Treatment",fill="Treatment")print(px)})# Assembling graphicsfinal_plot <-wrap_plots(plots, ncol =4)+plot_layout(guides ="collect")+plot_annotation(tag_levels ='A',title="Element Use Efficiency. After 100 random drawing",subtitle="")&theme(legend.position ='bottom')png(here::here("report/iono/plot/boxplot_EUE_100rep_treatment.png"), width =29.7, height =42, units ='cm', res =900)final_plotdev.off()
11.2.3.2 For sREU
Code
# warning, concentration in µg/g en root in g. df_EUE_sREU=read.csv(here::here("data/iono/output/random_drawing_EUE_sREU_rep100.csv"))[-1] %>%#filter(sEUpE>0) %>% dplyr::rename(sREU=sEUpE)# Graphics creation and assemblyvector_compound=levels(as.factor(as.character(df_EUE_sREU$compound_r1)))plots <-lapply(1:length(vector_compound), function(i) { df_EUE_sREU_for_plettre<-df_EUE_sREU %>% dplyr::rename(condition=condition_r2) %>%mutate(genotype=ifelse(str_sub(condition, 1,3)=="Sto","Stocata","Wendy")) %>%mutate(water_condition=str_sub(condition,5,6)) %>%mutate(heat_condition=str_sub(condition,8,9)) %>%filter(compound_r1==vector_compound[i]) %>%mutate(condition=factor(condition,levels=c("Wen_WW_OT","Wen_WW_HS","Wen_WS_OT","Wen_WS_HS","Sto_WW_OT","Sto_WW_HS","Sto_WS_OT","Sto_WS_HS")))FigX_stat=stat_analyse(data=df_EUE_sREU_for_plettre %>%mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>%mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>%mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>%drop_na(climat_condition,sREU),column_value ="sREU",category_variables =c("climat_condition"),grp_var ="genotype",show_plot = T,outlier_show = F, label_outlier ="compile_rep",biologist_stats = T,Ylab_i =paste0("sREU (µg ",vector_compound[i],"[gBMroot day-1]-1)"), control_conditions =c("WW_OT"),strip_normale = F,hex_pallet = climate_pallet)px=FigX_stat[["plot"]] +theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major =element_blank(),panel.grid.minor =element_blank(),legend.position ="bottom",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+labs(color="Treatment",fill="Treatment")print(px)})# Assembling graphicsfinal_plot <-wrap_plots(plots, ncol =4)+plot_layout(guides ="collect")+plot_annotation(tag_levels ='A',title="Specific Root Element Uptake. After 100 random drawing",subtitle="")&theme(legend.position ='bottom')png(here::here("report/iono/plot/boxplot_sREU_100rep_treatment.png"), width =29.7, height =48, units ='cm', res =900)final_plotdev.off()
11.3 Efficiency calculated with the average of harvest 1
11.3.1 Function for calculate EUE & sREU
Function for Element Use Efficiency based on H1 mean (harvest 1, control plant) and plant at harvest 2 (control or also treated condition). Each time we combine the same genotype. Idem for specific Root Element Uptake but is an integration during between date and we use root weight.
---output: html_documenteditor_options: chunk_output_type: console---## Pre-processing for efficiency in ionomic analyse```{r setup,message=FALSE, warning=FALSE,echo=T}# pkglibrary(readxl)library(tidyverse)library(dplyr)options(dplyr.summarise.inform = FALSE)library(stringr)library(tictoc)library(patchwork)# srcsource(here::here("src/function/stat_function/stat_analysis_main.R")) # to make plot # cosmeticsclimate_pallet=read_excel(here::here("data/color_palette.xlsm")) %>% filter(set == "climat_condition") %>% dplyr::select(color, treatment) %>% pull(color) %>% setNames(read_excel(here::here("data/color_palette.xlsm")) %>% filter(set == "climat_condition") %>% pull(treatment) )```## Data importation```{r, eval=F}plant_info=read_excel(here::here("data/plant_information.xlsx"))df_ionomic=read.csv(here::here("data/iono/output/ionomic.csv"))df_ionomic %>% filter(condition=="Sto_WS_HS") %>% filter(compound=="Cd111")```::: callout-warning## Beware of powder assemblies for two plants of the same condition and outliers- Note that when I pooler I consider twice the same value for two different plants. Ex: 1067 and 1069 pooler. The result goes to 1067 and 1069 not to 1067&1069)- I also removed the two plants 1039 and 1087 on 14/12/2022 for random draws (two plants out of type).:::## Efficicence with bootstrap### Function for bootstrap EUE & sREUFunction for **Element Use Efficiency** based on random drawing between plant at harvest 1 (control plant) and plant at harvest 2 (control or also treated condition). Each time we combine the same genotype. Idem for **specific Root Element Uptake** but is an integration during between date and we use root weight.```{r, eval=F}# warning, concentration in µg/g en root in g. random_drawing_iono_EUE_sREU<-function(rep){ set.seed(1) df_compile=data.frame() #do it for each compound!!! #Do not take more than one organ because we are looking at the total quantity of an element df_ionomic_select=df_ionomic %>% filter(compartiment=="leaf") %>% drop_na(sum_by_component) %>% filter(compound!="Sb121") %>% filter(num_compartiment==3) %>% drop_na(weight_total) %>% filter(sum_by_component>0) %>% filter(!plant_num%in% c(1039,1087)) list_condition=c("Sto_WW_OT","Sto_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS") for(e in levels(as.factor(as.character(df_ionomic_select$compound)))){ tic() for(cond in list_condition){ for (i in 1:rep){ #for R2 df_r2=df_ionomic_select %>% filter(recolte=="2") %>% filter(compound==e)%>% filter(condition==cond) df_r2_taking=df_r2 %>% filter(plant_num==sample(df_r2$plant_num,1,replace=T)) %>% dplyr::select(weight_total,condition,compound,sum_by_component,plant_num,recolte,date_recolte,weight_root) #now for r1 if(str_sub(df_r2_taking$condition, 1, 3)=="Sto"){ df_r1=df_ionomic_select %>% filter(recolte=="1") %>% filter(compound==e) %>% filter(condition=="Sto_WW_OT") df_r1_taking=df_r1 %>% filter(plant_num==sample(df_r1$plant_num,1,replace=T)) %>% dplyr::select(weight_total,condition,compound,sum_by_component,plant_num,recolte,date_recolte,weight_root) }else { df_r1=df_ionomic_select %>% filter(recolte=="1") %>% filter(compound==e) %>% filter(condition=="Wen_WW_OT") df_r1_taking=df_r1 %>% filter(plant_num==sample(df_r1$plant_num,1,replace=T)) %>% dplyr::select(weight_total,condition,compound,sum_by_component,plant_num,recolte,date_recolte,weight_root) } colnames(df_r1_taking)=c("weight_total_r1" ,"condition_r1" ,"compound_r1" ,"sum_by_component_r1" ,"plant_num_r1","recolte_r1","date_recolte_r1","weight_root_r1") colnames(df_r2_taking)=c("weight_total_r2" ,"condition_r2" ,"compound_r2" ,"sum_by_component_r2" ,"plant_num_r2","recolte_r2","date_recolte_r2","weight_root_r2") df_bind=cbind(df_r2_taking,df_r1_taking) df_bind$rep=i ####################### Element Uptake Eficiency ########################## df_bind$EUE= (df_bind$weight_total_r2-df_bind$weight_total_r1) / (df_bind$sum_by_component_r2-df_bind$sum_by_component_r1) #juste to verify df_bind$EUE_numerator=(df_bind$weight_total_r2-df_bind$weight_total_r1) df_bind$EUE_denominator=(df_bind$sum_by_component_r2-df_bind$sum_by_component_r1) ####################### Specific Root Element Uptake ########################## df_bind$sEUpE=(df_bind$sum_by_component_r2-df_bind$sum_by_component_r1)/ (( (as.numeric(as.Date(df_bind$date_recolte_r2)-as.Date(df_bind$date_recolte_r1))) * (df_bind$weight_root_r2+df_bind$weight_root_r1)/2 )) df_compile=rbind(df_compile,df_bind) } print(paste(sep="_",e,cond))} toc() } df_compile$compile_rep=paste(sep="_",df_compile$rep,df_compile$condition_r2) #df_compile =df_compile %>% filter(EUE>0) #write result write.csv(df_compile,here::here(paste(sep="","data/iono/output/random_drawing_EUE_sREU", #as.character(format(Sys.time(), "%Y%m%d_%H%M%S")), "_rep",rep,".csv"))) return(df_compile)}```### Function execution with 100 repetitions::: callout-warning### This function can take a very long time !!!:::```{r, eval=F}rep=100df_EUE_sREU=random_drawing_iono_EUE_sREU(rep=rep) #function little bit slow but very usefull```### Graphical verification#### For EUE::: callout-warning## I don't know if it's right, but sometimes there are negative values.I removed them to generate the graphs:::```{r,eval=F}# Warning, concentration in µg/g en root in g. df_EUE_sREU=read.csv(here::here("data/iono/output/random_drawing_EUE_sREU_rep100.csv"))[-1] #%>% #filter(EUE>0)# Graphics creation and assemblyvector_compound=levels(as.factor(as.character(df_EUE_sREU$compound_r1)))plots <- lapply(1:length(vector_compound), function(i) { df_EUE_sREU_for_plettre<-df_EUE_sREU %>% dplyr::rename(condition=condition_r2) %>% mutate(genotype=ifelse(str_sub(condition, 1,3)=="Sto","Stocata","Wendy")) %>% mutate(water_condition=str_sub(condition,5,6)) %>% mutate(heat_condition=str_sub(condition,8,9)) %>% filter(compound_r1==vector_compound[i]) %>% mutate(condition=factor(condition,levels=c("Wen_WW_OT","Wen_WW_HS","Wen_WS_OT","Wen_WS_HS","Sto_WW_OT","Sto_WW_HS","Sto_WS_OT","Sto_WS_HS")))FigX_stat=stat_analyse( data=df_EUE_sREU_for_plettre %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,EUE), column_value = "EUE", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "compile_rep", biologist_stats = T, Ylab_i = paste0("EUE for ",vector_compound[i], " (g.µgElement-1)"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet)px=FigX_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "bottom",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+labs(color="Treatment",fill="Treatment")print(px)})# Assembling graphicsfinal_plot <- wrap_plots(plots, ncol = 4)+ plot_layout(guides = "collect")+ plot_annotation(tag_levels = 'A', title="Element Use Efficiency. After 100 random drawing", subtitle="")& theme(legend.position = 'bottom')png(here::here("report/iono/plot/boxplot_EUE_100rep_treatment.png"), width = 29.7, height = 42, units = 'cm', res = 900)final_plotdev.off()```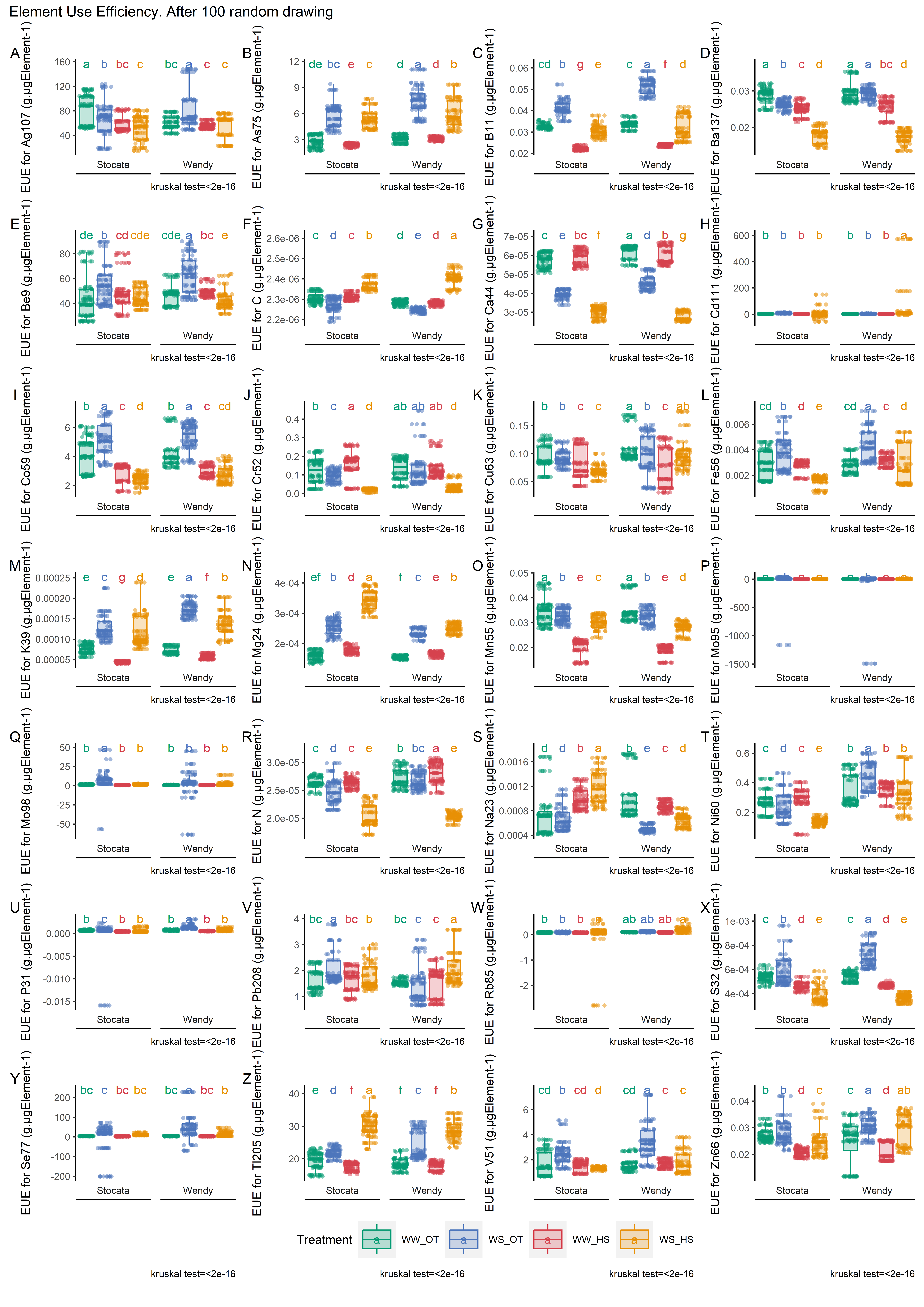#### For sREU```{r,eval=F}# warning, concentration in µg/g en root in g. df_EUE_sREU=read.csv(here::here("data/iono/output/random_drawing_EUE_sREU_rep100.csv"))[-1] %>% #filter(sEUpE>0) %>% dplyr::rename(sREU=sEUpE)# Graphics creation and assemblyvector_compound=levels(as.factor(as.character(df_EUE_sREU$compound_r1)))plots <- lapply(1:length(vector_compound), function(i) { df_EUE_sREU_for_plettre<-df_EUE_sREU %>% dplyr::rename(condition=condition_r2) %>% mutate(genotype=ifelse(str_sub(condition, 1,3)=="Sto","Stocata","Wendy")) %>% mutate(water_condition=str_sub(condition,5,6)) %>% mutate(heat_condition=str_sub(condition,8,9)) %>% filter(compound_r1==vector_compound[i]) %>% mutate(condition=factor(condition,levels=c("Wen_WW_OT","Wen_WW_HS","Wen_WS_OT","Wen_WS_HS","Sto_WW_OT","Sto_WW_HS","Sto_WS_OT","Sto_WS_HS")))FigX_stat=stat_analyse( data=df_EUE_sREU_for_plettre %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,sREU), column_value = "sREU", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "compile_rep", biologist_stats = T, Ylab_i = paste0("sREU (µg ",vector_compound[i],"[gBMroot day-1]-1)"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet)px=FigX_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "bottom",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+labs(color="Treatment",fill="Treatment")print(px)})# Assembling graphicsfinal_plot <- wrap_plots(plots, ncol = 4)+ plot_layout(guides = "collect")+ plot_annotation(tag_levels = 'A', title="Specific Root Element Uptake. After 100 random drawing", subtitle="")& theme(legend.position = 'bottom')png(here::here("report/iono/plot/boxplot_sREU_100rep_treatment.png"), width = 29.7, height = 48, units = 'cm', res = 900)final_plotdev.off()```## Efficiency calculated with the average of harvest 1### Function for calculate EUE & sREUFunction for **Element Use Efficiency** based on H1 mean (harvest 1, control plant) and plant at harvest 2 (control or also treated condition). Each time we combine the same genotype. Idem for **specific Root Element Uptake** but is an integration during between date and we use root weight.```{r}# warning, concentration in µg/g en root in g. function_iono_EUE_sREU<-function(){ df_compile=data.frame() df_ionomic_select=df_ionomic %>%filter(compartiment=="leaf") %>%drop_na(sum_by_component) %>%filter(compound!="Sb121") %>%filter(num_compartiment==3) %>%drop_na(weight_total) %>%filter(sum_by_component>0) %>%filter(!plant_num%in%c(1039,1087)) list_condition=c("Sto_WW_OT","Sto_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS")for(e inlevels(as.factor(as.character(df_ionomic_select$compound)))){for(cond in list_condition){ df_r2=df_ionomic_select %>%filter(recolte=="2") %>%filter(compound==e)%>%filter(condition==cond)for (i in1:nrow(df_r2)){#for R2 plant_num_i<-df_r2$plant_num[i] df_r2_taking=df_r2 %>%filter(plant_num==plant_num_i) %>% dplyr::select(condition, compound, recolte, date_recolte, plant_num,weight_total,weight_root,sum_by_component)#now for r1if(str_sub(df_r2_taking$condition, 1, 3)=="Sto"){ df_r1=df_ionomic_select %>%filter(recolte=="1") %>%filter(compound==e) %>%filter(condition=="Sto_WW_OT") df_r1_mean=df_r1 %>% dplyr::group_by(condition,compound,recolte,date_recolte)%>% dplyr::summarise(across(c("weight_total","weight_root", "sum_by_component"), ~mean(.x, na.rm =TRUE))) %>% dplyr::rename_all(~paste0(., "_H1")) }else { df_r1=df_ionomic_select %>%filter(recolte=="1") %>%filter(compound==e) %>%filter(condition=="Wen_WW_OT") df_r1_mean=df_r1 %>% dplyr::group_by(condition,compound,recolte,date_recolte)%>% dplyr::summarise(across(c("weight_total","weight_root", "sum_by_component"), ~mean(.x, na.rm =TRUE))) %>% dplyr::rename_all(~paste0(., "_H1")) }colnames(df_r1_mean)=c("condition_H1" ,"compound_H1" ,"Harvest_1","date_recolte_H1","weight_total_H1" ,"weight_root_H1","sum_by_component_H1")colnames(df_r2_taking)=c("condition_H2" ,"compound_H2", "Harvest_H2", "date_recolte_H2", "plant_num_H2","weight_total_H2", "weight_root_H2","sum_by_component_H2") df_bind=cbind(df_r2_taking,df_r1_mean)####################### Element Uptake Eficiency ########################## df_bind$EUE= (df_bind$weight_total_H2-df_bind$weight_total_H1) / (df_bind$sum_by_component_H2-df_bind$sum_by_component_H1)#juste to verify df_bind$EUE_numerator=(df_bind$weight_total_H2-df_bind$weight_total_H1) df_bind$EUE_denominator=(df_bind$sum_by_component_H2-df_bind$sum_by_component_H1)####################### Specific Root Element Uptake ########################## df_bind$sEUpE=(df_bind$sum_by_component_H2-df_bind$sum_by_component_H1)/ (( (as.numeric(as.Date(df_bind$date_recolte_H2)-as.Date(df_bind$date_recolte_H1))) * (df_bind$weight_root_H2+df_bind$weight_root_H1)/2)) df_compile=rbind(df_compile,df_bind) }cat(paste(e," ",cond, "\n"))} }#write resultwrite.csv(df_compile,here::here("data/iono/output/EUE_sREU_1mean.csv"))return(df_compile)}```### Function execution ```{r, eval=F}df_EUE_sREU=function_iono_EUE_sREU()```### Graphical verification#### For EUE::: callout-warning## I don't know if it's right, but sometimes there are negative values.I removed them to generate the graphs:::```{r,eval=F}# warning, concentration in µg/g en root in g. df_EUE_sREU=read.csv(here::here("data/iono/output/EUE_sREU_1mean.csv"))[-1]# %>% #filter(EUE>0)# Graphics creation and assemblyvector_compound=levels(as.factor(as.character(df_EUE_sREU$compound_H1)))plots <- lapply(1:length(vector_compound), function(i) {cat_col(c(vector_compound[i],"\n"),color = "green") df_EUE_sREU_for_plettre<-df_EUE_sREU %>% dplyr::rename(condition=condition_H2) %>% mutate(genotype=ifelse(str_sub(condition, 1,3)=="Sto","Stocata","Wendy")) %>% mutate(water_condition=str_sub(condition,5,6)) %>% mutate(heat_condition=str_sub(condition,8,9)) %>% filter(compound_H1==vector_compound[i]) %>% mutate(condition=factor(condition,levels=c("Wen_WW_OT","Wen_WW_HS","Wen_WS_OT","Wen_WS_HS","Sto_WW_OT","Sto_WW_HS","Sto_WS_OT","Sto_WS_HS")))FigX_stat=stat_analyse( data=df_EUE_sREU_for_plettre %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,EUE), column_value = "EUE", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = T, label_outlier = "plant_num_H2", biologist_stats = T, Ylab_i = paste0("EUE for ",vector_compound[i], " (g.µgElement-1)"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet)px=FigX_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "bottom",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+labs(color="Treatment",fill="Treatment")print(px)})# Assembling graphicsfinal_plot <- wrap_plots(plots, ncol = 4)+ plot_layout(guides = "collect")+ plot_annotation(tag_levels = 'A', title="Element Use Efficiency. Mean in harvest 1", subtitle="")& theme(legend.position = 'bottom')png(here::here("report/iono/plot/boxplot_EUE_1mean_treatment_outlier_show.png"), width = 29.7, height = 42, units = 'cm', res = 900)final_plotdev.off()```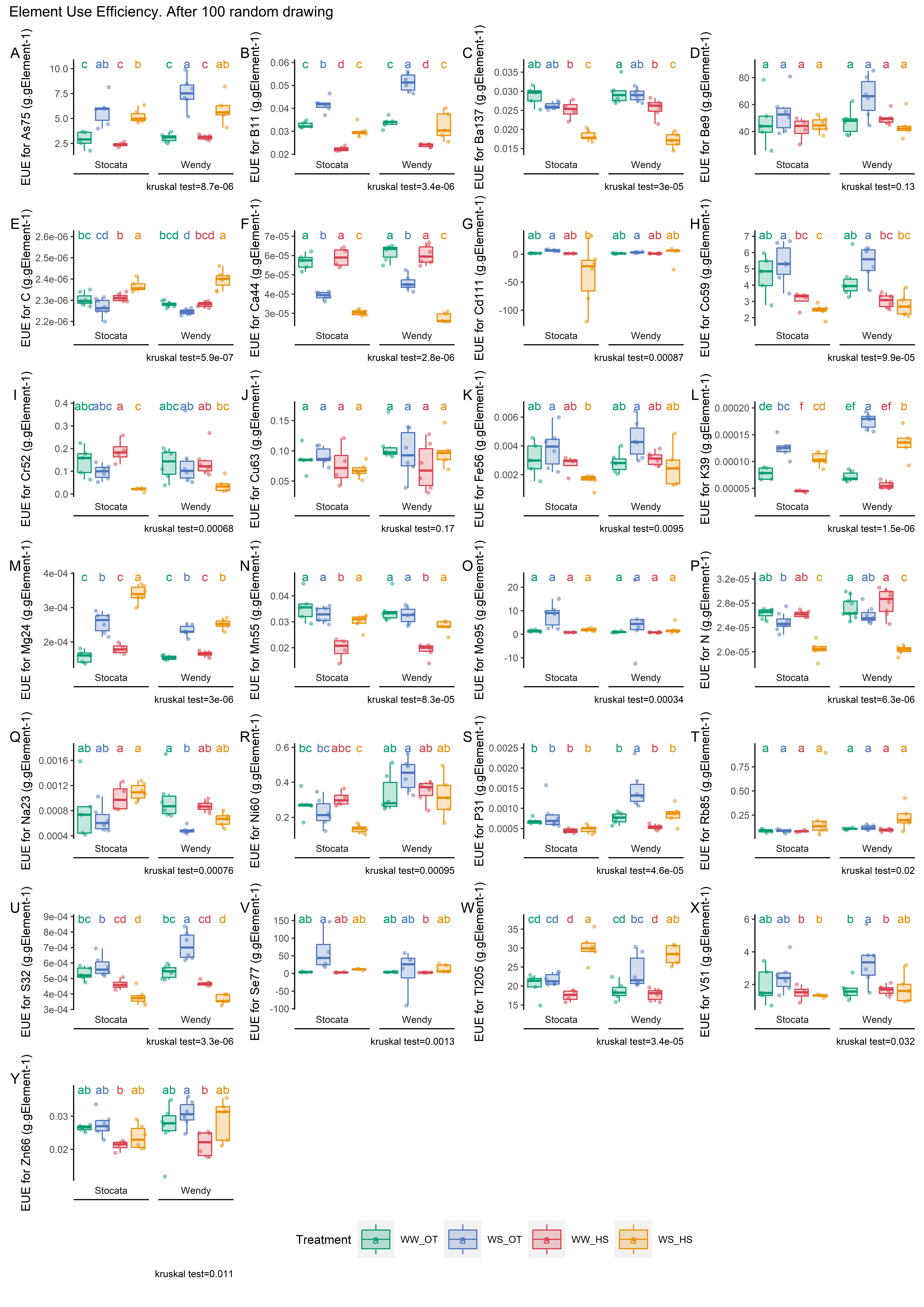#### For sREU```{r,eval=F}# warning, concentration in µg/g en root in g. df_EUE_sREU=read.csv(here::here("data/iono/output/EUE_sREU_1mean.csv"))[-1] %>% #filter(sEUpE>0) %>% dplyr::rename(sREU=sEUpE)# Graphics creation and assemblyvector_compound=levels(as.factor(as.character(df_EUE_sREU$compound_H1)))plots <- lapply(1:length(vector_compound), function(i) { cat_col(c(vector_compound[i],"\n"),color = "green") df_EUE_sREU_for_plettre<-df_EUE_sREU %>% dplyr::rename(condition=condition_H2) %>% mutate(genotype=ifelse(str_sub(condition, 1,3)=="Sto","Stocata","Wendy")) %>% mutate(water_condition=str_sub(condition,5,6)) %>% mutate(heat_condition=str_sub(condition,8,9)) %>% filter(compound_H1==vector_compound[i]) %>% mutate(condition=factor(condition,levels=c("Wen_WW_OT","Wen_WW_HS","Wen_WS_OT","Wen_WS_HS","Sto_WW_OT","Sto_WW_HS","Sto_WS_OT","Sto_WS_HS")))FigX_stat=stat_analyse( data=df_EUE_sREU_for_plettre %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,sREU), column_value = "sREU", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = T, label_outlier = "plant_num_H2", biologist_stats = T, Ylab_i = paste0("sREU (µg ",vector_compound[i],"[gBMroot day-1]-1)"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet)px=FigX_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "bottom",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+labs(color="Treatment",fill="Treatment")print(px)})# Assembling graphicsfinal_plot <- wrap_plots(plots, ncol = 4)+ plot_layout(guides = "collect")+ plot_annotation(tag_levels = 'A', title="Specific Root Element Uptake. Mean in Harvest 1", subtitle="")& theme(legend.position = 'bottom')png(here::here("report/iono/plot/boxplot_sREU_1mean_treatment_outlier_show.png"), width = 29.7, height = 48, units = 'cm', res = 900)final_plotdev.off()```