--- output: html_document editor_options: chunk_output_type: console --- ## Rapid analysis of root architecture data ```{r setup,message=FALSE, warning=FALSE,echo=T} #package library(tidyr) library(readxl) library(ggplot2) library(plotly) library(vroom) library(ggExtra) library(gridExtra) library(ggpubr) library(cowplot) # for plotgrid library(stringr) library(dplyr) library(gt) # gramar table library(DT) library(tidyverse) library(here) library(png) # just to put the png as background in the figure library(patchwork) library(psych) # for correlation library(PerformanceAnalytics) #for correlation #src source(here::here("src/function/graph_function.R")) source(here::here("src/function/data_importation_xp1.R")) source(here::here("src/function/stat_function/stat_analysis_main.R")) # for make plot #source(here::here("xp1_analyse/script/function/top_cor_function.R")) #for top correlation and show plot of correlation set.seed(1) ``` ## Data importation ```{r,eval=T, message=F, results='hide'} coo_root_tips_select=read.csv(here::here("data/root/output/coo_root_tips_select.csv"))%>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Sto_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) coo_root_angle_select=read.csv(here::here("data/root/output/coo_root_angle_select.csv"))%>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Sto_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) df_global_archi=import_kinetic(df_ra_i = T,df_height_plant_i = T,df_root_tips_nb_10000_i = T,df_root_tips_angle_i = T,df_root_tips_length_mean_i = T, df_root_tips_length_sum_i = T, df_root_nb_ramif_i = T,df_root_tips_nb_macro_i = T,key = "plant_num_shooting_date") %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Sto_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% filter(days_after_transplantation!=3) #all data compile by taskid, shooting date and plant num # cosmetics climate_pallet=read_excel(here::here("data/color_palette.xlsm")) %>% filter(set == "climat_condition") %>% dplyr::select(color, treatment) %>% pull(color) %>% setNames(read_excel(here::here("data/color_palette.xlsm")) %>% filter(set == "climat_condition") %>% pull(treatment) ) ``` ## Analyse of kinetics data made with deep learning ```{r,eval=F} # Definition of variables variable_archi=c("root","area","largeur","density","profondeur","length_skull","diff_correct_plant_height","surface", "volume") legende_variable_archi=c("Root projected area (cm²)","Area of the convexhull (cm²)","Width of the root system (cm)","Density of the root system","Depth of the root system (cm)","Length total of the root system (cm)","Height of the plant (cm)", "Root surface area (cm²)", "Volume of the root (cm^3)") # Data generation df_global_archi_n2=df_global_archi %>% filter(climat_condition!="NA_NA") %>% group_by(climat_condition,days_after_transplantation) %>% summarise_at(.vars = variable_archi, .funs = c(mean="mean", sd="sd"),na.rm=T) df_global_archi_n=df_global_archi %>% filter(climat_condition!="NA_NA") %>% group_by(climat_condition,days_after_transplantation,plant_num) %>% summarise_at(.vars = variable_archi, .funs = c(mean="mean", sd="sd"),na.rm=T) # Graphics creation and assembly plots <- lapply(1:length(variable_archi), function(i) { i_mean=paste0(variable_archi[i],"_mean") min_y <- min(df_global_archi_n2[,i_mean]) px=df_global_archi_n2 %>% ggplot(aes_string(x="days_after_transplantation",y=i_mean,col="climat_condition",fill="climat_condition"))+ geom_smooth(data=df_global_archi_n,aes_string(x="days_after_transplantation",y=i_mean,color="climat_condition"),color = "transparent",alpha=0.25)+ geom_line(size=1,se=T)+ geom_point()+ scale_x_continuous(breaks=seq(0, 25, 2))+ ylab(paste0(legende_variable_archi[i]))+ xlab("Number of days after transplantation")+ scale_color_manual(values=climate_pallet)+ theme_bw()+ theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())+ scale_fill_manual(values=climate_pallet)+labs(color="Treatment",fill="Treatment") #plotly::ggplotly(px) print(px) }) # Assembling graphics final_plot <- wrap_plots(plots, ncol = 2)+ plot_layout(guides = "collect")+ guide_area()+ plot_annotation(tag_levels = 'A', title="Combined graph of kinetic variables measured during the experiment", subtitle="Comparison between treatments without taking genotype into account") final_plot ggsave(here::here("report/root_architecture/plot/kinetic_climat_condition.svg"), final_plot, height = 12,width = 7) # By condition (for the two genotype) df_global_archi_n2=df_global_archi %>% group_by(condition,climat_condition,genotype,days_after_transplantation) %>% summarise_at(.vars = variable_archi, .funs = c(mean="mean", sd="sd"),na.rm=T) df_global_archi_n=df_global_archi %>% group_by(condition,climat_condition,genotype,days_after_transplantation,plant_num) %>% summarise_at(.vars = variable_archi, .funs = c(mean="mean", sd="sd"),na.rm=T) #dev.off() plots <- lapply(1:length(variable_archi), function(i) { i_mean=paste0(variable_archi[i],"_mean") px=df_global_archi_n2 %>% drop_na(climat_condition) %>% ggplot(aes_string(x="days_after_transplantation",y=i_mean,col="climat_condition",fill="climat_condition",linetype="genotype"))+ geom_smooth(data=df_global_archi_n,aes_string(x="days_after_transplantation",y=i_mean,color="climat_condition"),color = "transparent",alpha=0.25)+ geom_line(size=1,se=T)+geom_point()+ scale_x_continuous(breaks=seq(0, 25, 2))+ ylab(paste0(legende_variable_archi[i]))+xlab("Number of days after transplantation")+ scale_color_manual(values=climate_pallet)+ theme_bw()+ theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())+ scale_fill_manual(values=climate_pallet)+ labs(color="Treatment",fill="Treatment",linetype="Genotype") #plotly::ggplotly(px) print(px) }) # Assembling graphics final_plot <- wrap_plots(plots, ncol = 2)+ plot_layout(guides = "collect")+ guide_area()+ plot_annotation(tag_levels = 'A', title="Combined graph of kinetic variables measured during the experiment", subtitle="Comparison between treatments and genotype") final_plot ggsave(here::here("report/root_architecture/plot/kinetic_condition.svg"), final_plot, height = 14,width = 7) #stats for the root surface projected nb_day=20 df_global_archi_select=df_global_archi %>% filter(days_after_transplantation==nb_day) # # Fig_root_arch=Plettre_grp( # Z=df_global_archi_select, # X=df_global_archi_select$condition, # Y=df_global_archi_select$root, # Xlab = "Condition", # Ylab = paste0("Root projected area (cm²)"), # Y_bis = "root", # Tukey=T, # outlier_show =F , # label_outlier = "plant_num", # export_png = T,export_path = here::here(paste0("report/root_architecture/plot/root_projected_area_with_label_plant_num_",nb_day,"DAT",".png")),export_width = 17, export_height = 15, # legend_del = F, # title_plot = paste0(nb_day," days after transplantation"), # print_stats = F # ) plot_x=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,length_skull), column_value = "surface", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0("Root surface area (cm²)"), control_conditions = c("WW_OT"), strip_normale = T, hex_pallet = climate_pallet ) ggsave(here::here("report/root_architecture/plot/root_surface_area_20DAT.svg"), plot_x[["plot"]], height = 4,width = 5) # #calcul for now the decrease at 9 days after transplantation # df_global_archi_select %>% dplyr::group_by(heat_condition,genotype) %>% # dplyr::summarise(mean_area=mean(root)) %>% as.data.frame() # # df_global_archi_select %>% dplyr::group_by(heat_condition,genotype) %>% # #filter(genotype=="Stocata") %>% # filter(climat_condition %in% c("WW_OT","WW_HS")) %>% # dplyr::summarise(mean_area=mean(root)) %>% as.data.frame() %>% mutate(variation_rate=(mean_area[heat_condition=="HS"]-mean_area[heat_condition=="OT"])*100/(mean_area[heat_condition=="OT"])) %>% filter(heat_condition=="HS") # # # list_days_after_transplantation=levels(as.factor(df_global_archi %>% pull(days_after_transplantation))) # # #student pour savoir quand le stress hydrique est significativement diférent par rapport au controle # df_global_archi %>% filter(days_after_transplantation==17) %>% # group_by(genotype) %>% # summarise_each(funs(t.test(.[climat_condition == "WW_OT"], .[climat_condition == "WS_OT"])$p.value), vars = root) # # # df_global_archi %>% filter(days_after_transplantation==19) %>% # #group_by(genotype) %>% # summarise_each(funs(t.test(.[condition == "Sto_WS_OT"], .[condition == "Wen_WS_OT"])$p.value), vars = root) # # # df_global_archi %>% # filter(days_after_transplantation>4) %>% # group_by(genotype,days_after_transplantation) %>% # summarise_each(funs(t.test(.[climat_condition == "WW_OT"], .[climat_condition == "WS_OT"])$p.value), vars =diff_correct_plant_height) # # df_global_archi %>% # filter(days_after_transplantation>4) %>% # #group_by(genotype,days_after_transplantation) %>% # group_by(days_after_transplantation) %>% # summarise_each(funs(t.test(.[condition == "Sto_WW_HS"], .[condition == "Wen_WW_HS"])$p.value), vars =diff_correct_plant_height) # # df_global_archi %>% # filter(days_after_transplantation==20) %>% dplyr::group_by(condition) %>% # #filter(genotype=="Stocata") %>% # drop_na(diff_correct_plant_height) %>% # #filter(climat_condition %in% c("WW_OT","WW_HS")) %>% # dplyr::summarise(mean_height_plant=mean(diff_correct_plant_height)) %>% as.data.frame() %>% mutate(diff=mean_height_plant[condition=="Sto_WW_OT"]-mean_height_plant[condition=="Wen_WW_OT"]) # # # M1=lm(data = df_global_archi,root~genotype+days_after_transplantation+climat_condition) # summary(M1) ```   ```{r,eval=F} #only root projected area and height of the plant df_global_archi_select=df_global_archi %>% filter(days_after_transplantation==20) #%>% drop_na() i_mean=paste0(variable_archi[1],"_mean") p_root_projected=df_global_archi_n2 %>% drop_na(climat_condition) %>% ggplot(aes_string(x="days_after_transplantation",y=i_mean,col="climat_condition",fill="climat_condition",linetype="genotype"))+ geom_rect(xmin = 5, xmax = 20, ymin = -Inf, ymax = 0, fill = "#c9d5ea", alpha = 0.5,col="gray") + geom_rect(xmin = 5, xmax = 10, ymin = 0, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + geom_rect(xmin = 15, xmax = 20, ymin = 0, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + # geom_rect(aes(xmin=15,xmax = 18, ymin = -Inf,ymax = Inf), fill = 'gray80', alpha = 0.05,color=NA)+ # geom_rect(aes(xmin=5,xmax = 8, ymin = -Inf,ymax = Inf), fill = 'gray80', alpha = 0.05,color=NA)+ # geom_rect(aes(xmin=5,xmax = Inf, ymin = -Inf,ymax = 5), fill = '#5E5A93', alpha = 0.01,color=NA)+ # # geom_smooth(data=df_global_archi_n,aes_string(x="days_after_transplantation",y=i_mean,color="climat_condition"),color = "transparent",alpha=0.25)+ geom_line(size=1,se=T)+ geom_point()+ scale_x_continuous(breaks=seq(0, 25, 2))+ ylab(paste0(legende_variable_archi[8]))+xlab("Number of days after transplantation")+ theme_bw()+ theme( # Hide panel borders and remove grid lines panel.border = element_blank(), panel.grid.major = element_blank(), panel.grid.minor = element_blank(), # Change axis line axis.line = element_line(colour = "black") )+ scale_color_manual(values=climate_pallet)+ scale_fill_manual(values=climate_pallet)+ theme(legend.position = "right") + labs(color="Treatment",fill="Treatment",linetype="Genotype");p_root_projected i_mean=paste0(variable_archi[8],"_mean") p_root_surface=df_global_archi_n2 %>% drop_na(climat_condition) %>% ggplot(aes_string(x="days_after_transplantation",y=i_mean,col="climat_condition",fill="climat_condition",linetype="genotype"))+ geom_rect(xmin = 5, xmax = 20, ymin = -Inf, ymax = 0, fill = "#c9d5ea", alpha = 0.5,col="gray") + geom_rect(xmin = 5, xmax = 10, ymin = 0, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + geom_rect(xmin = 15, xmax = 20, ymin = 0, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + # geom_rect(aes(xmin=15,xmax = 18, ymin = -Inf,ymax = Inf), fill = 'gray80', alpha = 0.05,color=NA)+ # geom_rect(aes(xmin=5,xmax = 8, ymin = -Inf,ymax = Inf), fill = 'gray80', alpha = 0.05,color=NA)+ # geom_rect(aes(xmin=5,xmax = Inf, ymin = -Inf,ymax = 5), fill = '#5E5A93', alpha = 0.01,color=NA)+ # # geom_smooth(data=df_global_archi_n,aes_string(x="days_after_transplantation",y=i_mean,color="climat_condition"),color = "transparent",alpha=0.25)+ geom_line(size=1,se=T)+ geom_point()+ scale_x_continuous(breaks=seq(0, 25, 2))+ ylab(paste0(legende_variable_archi[8]))+xlab("Number of days after transplantation")+ theme_bw()+ theme( # Hide panel borders and remove grid lines panel.border = element_blank(), panel.grid.major = element_blank(), panel.grid.minor = element_blank(), # Change axis line axis.line = element_line(colour = "black") )+ scale_color_manual(values=climate_pallet)+ scale_fill_manual(values=climate_pallet)+ theme(legend.position = "right") + labs(color="Treatment",fill="Treatment",linetype="Genotype");p_root_projected # #export only root # svg(here::here("xp1_analyse/plot/root_architecture/root_projected_area_poster.svg"), width = 15/2.5, height = 10/2.5)#, units = 'cm', res = 900) # grid.arrange (p_root_projected) # Make plot # dev.off() Fig1A_stat_surface=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,surface), column_value = "surface", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0(paste0("Root surface area (cm²)")), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig1A_stat_surface=Fig1A_stat_surface[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),legend.position = "none",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))#,plot.caption = element_text(vjust = 10)) #Fig1A_stat i_mean=paste0(variable_archi[7],"_mean") p_height_plant=df_global_archi_n2 %>% drop_na(climat_condition) %>% ggplot(aes_string(x="days_after_transplantation",y=i_mean,col="climat_condition",fill="climat_condition",linetype="genotype"))+ geom_rect(xmin = 5, xmax = 20, ymin = -Inf, ymax = 10, fill = "#c9d5ea", alpha = 0.5,col="gray") + geom_rect(xmin = 5, xmax = 10, ymin = 10, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + geom_rect(xmin = 15, xmax = 20, ymin = 10, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + geom_smooth(data=df_global_archi_n,aes_string(x="days_after_transplantation",y=i_mean,color="climat_condition"),color = "transparent",alpha=0.25)+ geom_line(size=1,se=T)+geom_point()+ scale_x_continuous(breaks=seq(0, 25, 2))+ ylab(paste0(legende_variable_archi[7]))+xlab("Number of days after transplantation")+ theme_bw()+ my_theme+ scale_color_manual(values=climate_pallet)+ scale_fill_manual(values=climate_pallet)+ theme(legend.position = "bottom") df_global_archi_select=df_global_archi %>% filter(days_after_transplantation==20) %>% drop_na(diff_correct_plant_height) Fig1B_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,diff_correct_plant_height), column_value = "diff_correct_plant_height", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0(paste0(legende_variable_archi[7])), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig1B_stat=Fig1B_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))#,plot.caption = element_text(vjust = 10)) Fig1=ggarrange(p_root_projected,Fig1A_stat,p_height_plant,Fig1B_stat,widths = c(1, 0.6),ncol=2, nrow=2,common.legend = TRUE, legend="bottom",labels = c("A","B","C","D"),align='h') Fig1 #export to delet #ggsave(here::here("xp1_analyse/plot/root_architecture/resum_Fig1.svg"), Fig1, width = 25, height = 25, units = "cm") png(here::here("report/root_architecture/plot/resum_Fig1.png"), width = 25, height = 25, units = 'cm', res = 900) grid.arrange (Fig1) # Make plot dev.off() png(here::here("report/root_architecture/plot/resum_Fig1_root.png"), width = 18, height = 12, units = 'cm', res = 900) ggarrange(p_root_projected, Fig1A_stat, widths = c(1, 0.6),ncol=2, nrow=1,common.legend = TRUE, legend="bottom",labels = c("A","B"),align='h') # Make plot dev.off() svg(here::here("report/root_architecture/plot/resum_Fig1_root.svg"), width = 7, height = 4) ggarrange(p_root_projected, Fig1A_stat, widths = c(1, 0.6),ncol=2, nrow=1,common.legend = TRUE, legend="bottom",labels = c("A","B"),align='h') # Make plot dev.off() svg(here::here("report/root_architecture/plot/resum_Fig1_root2.svg"), width = 7, height = 4) ggarrange(p_root_surface, Fig1A_stat_surface, widths = c(1, 0.6),ncol=2, nrow=1,common.legend = TRUE, legend="bottom",labels = c("A","B"),align='h') # Make plot dev.off() ``` 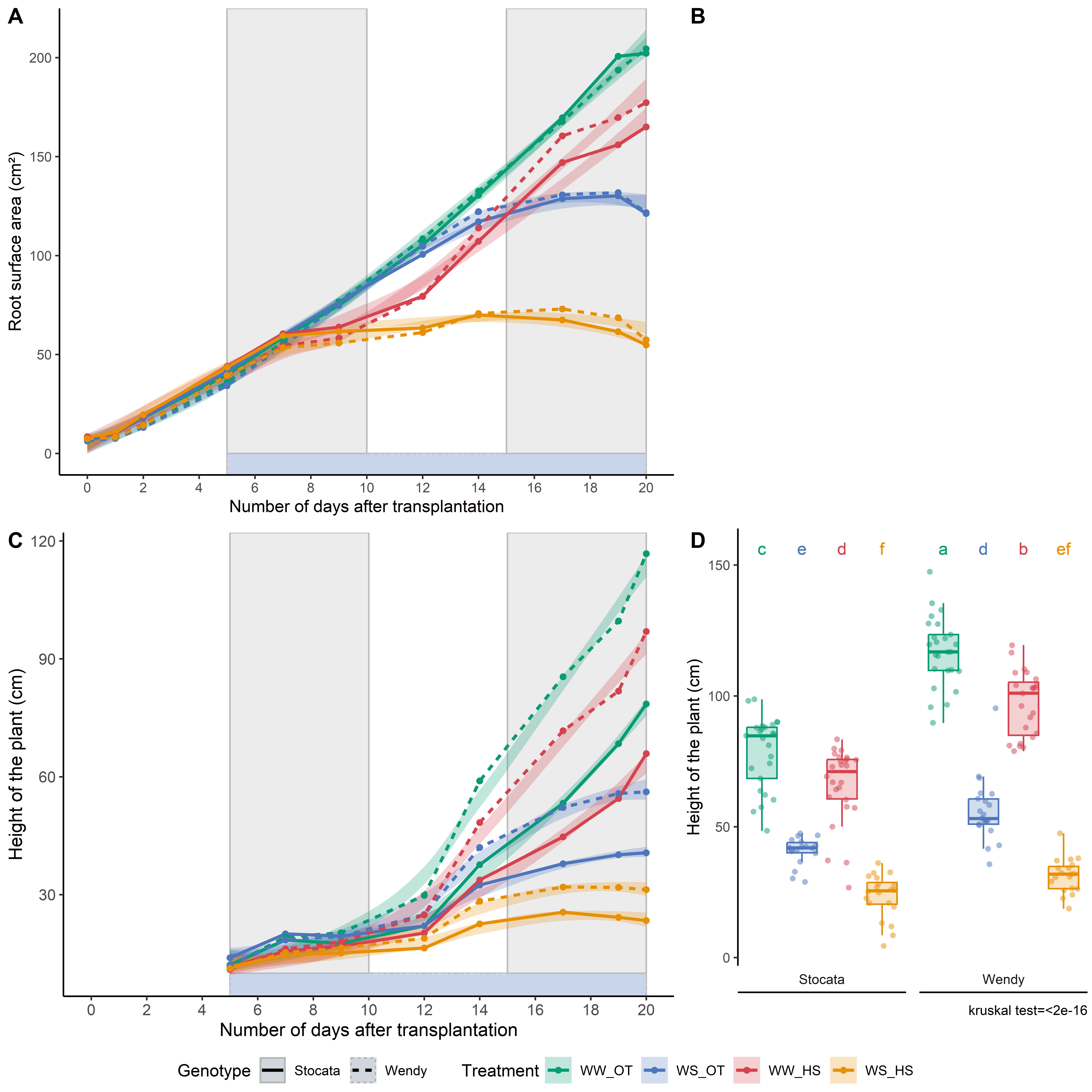 ## The speed of root growth ```{r, eval=FALSE} df_kinetic<-import_kinetic(df_height_plant_i = T, df_ra_i = T,key = "plant_num_shooting_date",date_shooting = "all") %>% filter(recolte==2) %>% dplyr::select("plant_num","condition","heat_condition", "water_condition","genotype", "length_skull", "diff_correct_plant_height", "climat_condition", "days_after_transplantation") %>% drop_na(length_skull) vec_plant_num<-levels(as.factor(as.character(df_kinetic$plant_num))) df_kinetic$velocity_length_root<- NA df_kinetic$velocity_height_plant<- NA df_kinetic2=data.frame() for(plant_num_x in vec_plant_num){ df_kinetic_x<-df_kinetic %>% filter(plant_num==plant_num_x) %>% arrange(days_after_transplantation) if(nrow(df_kinetic_x)<2){next} for (i in 1:nrow(df_kinetic_x)){ if(i==1){ df_kinetic_x$velocity_length_root[i]<-0 df_kinetic_x$velocity_height_plant[i]<-0 }else{ df_kinetic_x$velocity_length_root[i]<-(df_kinetic_x$length_skull[i]-df_kinetic_x$length_skull[i-1])/as.numeric(df_kinetic_x$days_after_transplantation[i]-df_kinetic_x$days_after_transplantation[i-1]) df_kinetic_x$velocity_height_plant[i]<-(df_kinetic_x$diff_correct_plant_height[i]-df_kinetic_x$diff_correct_plant_height[i-1])/as.numeric(df_kinetic_x$days_after_transplantation[i]-df_kinetic_x$days_after_transplantation[i-1]) } } df_kinetic2<-rbind(df_kinetic2,df_kinetic_x) } p_velocity_height_plant<- ggplot(df_kinetic2 %>% filter(velocity_height_plant>-5) %>% filter( velocity_height_plant<25),aes_string(x="days_after_transplantation",y="velocity_height_plant",col="climat_condition", fill="climat_condition",linetype="genotype"))+ geom_rect(xmin = 5, xmax = 20, ymin = -Inf, ymax = -2, fill = "#c9d5ea", alpha = 0.5,col="gray") + geom_rect(xmin = 5, xmax = 10, ymin = -2, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + geom_rect(xmin = 15, xmax = 20, ymin = -2, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray")+ geom_point(alpha=0.25)+ facet_grid(.~climat_condition)+ geom_smooth(data=df_kinetic2 %>% filter(velocity_height_plant>-5) %>% filter( velocity_height_plant<25), aes_string(x="days_after_transplantation",y="velocity_height_plant",color="climat_condition"),alpha=0.25)+ labs(fill="Treatment",color="Treatment",linetype="Genotype",y="Velocity of the height of the plant (cm/days)", x="Number of days after transplantation")+ theme_bw()+ #my_theme+ scale_color_manual(values=climate_pallet)+ scale_fill_manual(values=climate_pallet)+ theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), strip.text = element_blank(), legend.position = "bottom") p_velocity_length_root<- ggplot(df_kinetic2 %>% filter(velocity_length_root>-50),aes_string(x="days_after_transplantation",y="velocity_length_root",col="climat_condition", fill="climat_condition",linetype="genotype"))+ geom_rect(xmin = 5, xmax = 20, ymin = -Inf, ymax = -40, fill = "#c9d5ea", alpha = 0.5,col="gray") + geom_rect(xmin = 5, xmax = 10, ymin = -40, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray") + geom_rect(xmin = 15, xmax = 20, ymin = -40, ymax = Inf, fill = "lightgray", alpha = 0.02,col="gray")+ geom_point(alpha=0.25)+ facet_grid(.~climat_condition)+ geom_smooth(data=df_kinetic2 %>% filter(velocity_length_root>-50), aes_string(x="days_after_transplantation",y="velocity_length_root",color="climat_condition"),alpha=0.25)+ labs(fill="Treatment",color="Treatment",linetype="Genotype",y="Velocity root length (cm/days)", x="Number of days after transplantation")+ theme_bw()+ #my_theme+ scale_color_manual(values=climate_pallet)+ scale_fill_manual(values=climate_pallet)+ theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(), strip.text = element_blank(), legend.position = "bottom") # Compile the two plots svg(here::here("report/root_architecture/plot/velocity_height_plant_root_length.svg"), width = 10, height = 8) ggarrange(p_velocity_height_plant, p_velocity_length_root,ncol=1, nrow=2,common.legend = TRUE, legend="bottom",labels = c("A","B"),align='v') dev.off() png(here::here("report/root_architecture/plot/velocity_height_plant_root_length.png"), width = 10, height = 8, units = "in", res = 300) ggarrange(p_velocity_height_plant, p_velocity_length_root, ncol = 1, nrow = 2, common.legend = TRUE, legend = "bottom", labels = c("A", "B"), align = 'v') dev.off() ``` 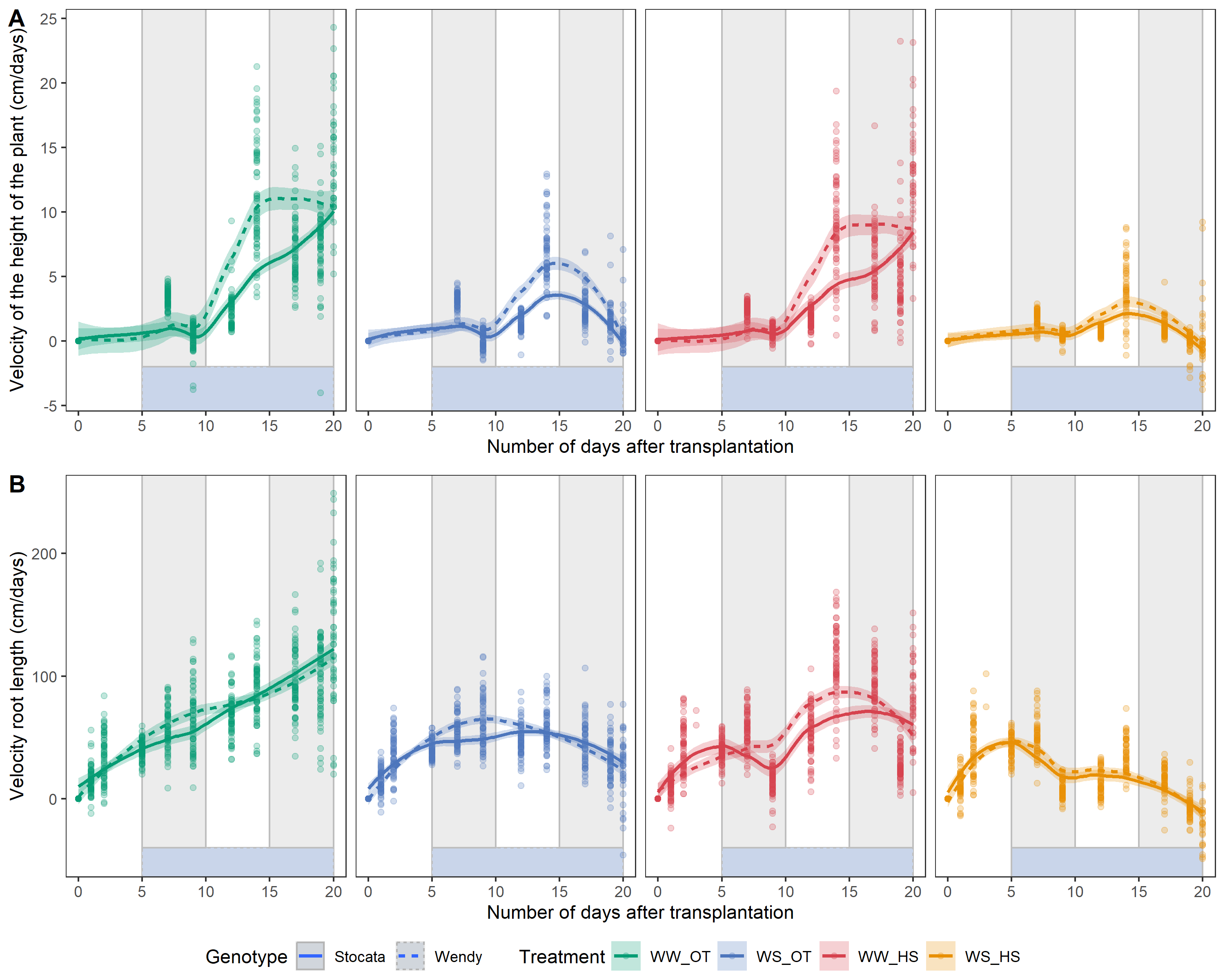 ## Angles and root tips  ### Number of root tips by branching ```{r, eval=F} #only root projected area and height of the plant df_global_archi_select=df_global_archi %>% filter(days_after_transplantation==19) %>% filter(plant_num!=116)#%>% drop_na() Fig1A_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,nb_tips_macro_1), column_value = "nb_tips_macro_1", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0("Number of root tips \n for branching 1"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig1A_stat=Fig1A_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+ labs(color="Treatment",fill="Treatment") Fig2A_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,nb_tips_macro_2), column_value = "nb_tips_macro_2", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0("Number of root tips \n for branching 2"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig2A_stat=Fig2A_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+ labs(color="Treatment",fill="Treatment") Fig3A_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,nb_tips_macro_3), column_value = "nb_tips_macro_3", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0("Number of root tips \n for branching 3"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig3A_stat=Fig3A_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+ labs(color="Treatment",fill="Treatment") Fig4A_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,sum_tips_macro), column_value = "sum_tips_macro", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = paste0("Number of root tips \n for all the plant"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig4A_stat=Fig4A_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none",plot.margin=unit(c(0.1,0.1,0.7,0.1), "cm"))+ labs(color="Treatment",fill="Treatment") FigNbTips=ggarrange(Fig1A_stat,Fig2A_stat,Fig3A_stat,Fig4A_stat ,ncol=2, nrow=2,common.legend = TRUE, legend="bottom",labels = c("A","B","C","D"),align='v') FigNbTips ``` ```{r,eval=FALSE} #################### If I compile the number of root tips into a single graph ################# # Interesting to look at the proportions for the number of root tips? df_global_archi_select=df_global_archi %>% filter(days_after_transplantation==19) #%>% drop_na() df_global_archi_select$condition<- dplyr::recode(df_global_archi_select$condition, Sto_WW_OT="Stocata_WW_OT", Sto_WS_OT="Stocata_WS_OT", Sto_WW_HS="Stocata_WW_HS", Sto_WS_HS="Stocata_WS_HS", Wen_WW_OT="Wendy_WW_OT", Wen_WS_OT="Wendy_WS_OT", Wen_WW_HS="Wendy_WW_HS", Wen_WS_HS="Wendy_WS_HS" ) #data_biomass=data_biomass %>% mutate(moda=factor(moda,levels=c("WW.OT","WS.OT","WW.HS","WS.HS"))) # df_global_archi_select$climat_condition<- dplyr::recode(df_global_archi_select$climat_condition, # WW_OT="WW.OT", # WS_OT="WS.OT", # WW_HS="WW.HS", # WS_HS="WS.HS" # ) #moyenne df_global_archi_select2=df_global_archi_select%>% # filter(!plante_code_simple %in% c(liste_error))%>% select(condition,climat_condition,genotype,nb_tips_macro_1,nb_tips_macro_2, nb_tips_macro_3,sum_tips_macro)%>%na.omit() mean_df_global_archi_select2=df_global_archi_select2 %>% # Specify data frame group_by(condition,climat_condition,genotype) %>% # Specify group indicator summarise_at(vars(nb_tips_macro_1,nb_tips_macro_2, nb_tips_macro_3), # Specify column list(mean)) # Specify function mean_df_global_archi_select2_bis=as.data.frame(mean_df_global_archi_select2) # add an adjuste column for eror bar for (i in 1:length(mean_df_global_archi_select2$condition)){ mean_df_global_archi_select2_bis$adj_macro3[i]=mean_df_global_archi_select2_bis$nb_tips_macro_1[i]+mean_df_global_archi_select2_bis$nb_tips_macro_2[i]+mean_df_global_archi_select2_bis$nb_tips_macro_3[i] mean_df_global_archi_select2_bis$adj_macro2[i]=mean_df_global_archi_select2_bis$nb_tips_macro_1[i]+mean_df_global_archi_select2_bis$nb_tips_macro_2[i] mean_df_global_archi_select2_bis$adj_macro1[i]=mean_df_global_archi_select2_bis$nb_tips_macro_1[i] } nb_tips_macro_vertical_mean <- mean_df_global_archi_select2 %>% pivot_longer(c(nb_tips_macro_1,nb_tips_macro_2, nb_tips_macro_3), names_to="Branching", values_to="nb_tips_mean") nb_tips_macro_vertical_mean_bis <- mean_df_global_archi_select2_bis %>% pivot_longer(c(adj_macro3,adj_macro2,adj_macro1), names_to="Branching", values_to="adj_mean") data_vertical_mean=cbind(nb_tips_macro_vertical_mean,nb_tips_macro_vertical_mean_bis[,8]) # addition of standard deviation sd_archi_macro=df_global_archi_select2 %>% # Specify data frame group_by(condition,climat_condition,genotype) %>% # Specify group indicator summarise_at(vars(nb_tips_macro_1,nb_tips_macro_2, nb_tips_macro_3), # Specify column list(sd)) # Specify function data_vertical_sd <- sd_archi_macro %>% pivot_longer(c(nb_tips_macro_1,nb_tips_macro_2, nb_tips_macro_3), names_to="Branching", values_to="tips_sd") data_resum=cbind(data_vertical_mean,data_vertical_sd$tips_sd) colnames(data_resum)=c("condition","climat_condition","genotype","Branching","nb_tips_mean","adj_mean","tips_sd") # change variables data_resum$Branching=dplyr::recode(data_resum$Branching, nb_tips_macro_1 = "Branching 1",nb_tips_macro_2= "Branching 2", nb_tips_macro_3 = "Branching 3") # graph #Fig1---- data_resum$Branching <- fct_relevel(data_resum$Branching, c("Branching 3", "Branching 2", "Branching 1" )) # Add a column that adjusts my error bars # add below the sd the letter data_resum$letter=c("cde","a","b", "ef","c","bc", "def","bc","bc", "f","d","c", "ab","ab","a", "a","c","bc", "abc","ab","bc", "bcd","d","c" ) # creation of a table for total plant stats mean_letter1=data_resum %>% # Specify data frame group_by(condition,climat_condition,genotype) %>% # Specify group indicator summarise_at(vars(nb_tips_mean), # Specify column list(sum)) mean_letter2=data_resum %>% # Specify data frame group_by(condition,climat_condition,genotype) %>% filter(Branching=="Branching 3")%>%select(tips_sd) mean_letter1=cbind(mean_letter1,mean_letter2[,4]) mean_letter1$adj_tot=mean_letter1$nb_tips_mean+mean_letter1$tips_sd # for letter Plettre_modif(Z=data_biomass_select,Y=data_biomass_select$Node_weight+data_biomass_select$Root_weight+data_biomass_select$PA,X=data_biomass_select$Genotype,Xlab="x",Ylab = "y") mean_letter1$letter=c("ab","de","cd","f","a","cd","bc","ef") color_branching <- c("#ffdbac","#e0ac69","#c68642") # graph biomass Fig_nb_tips=ggplot(data_resum, aes(x = climat_condition,y=nb_tips_mean,fill=Branching))+ geom_text(position = position_stack(vjust = 1.2),data = mean_letter1, aes(y = adj_tot, label = letter, fill = "Branching 1"),fontface = "bold",color="#000000")+ geom_bar(stat = "identity",color="black")+ theme_bw()+ geom_errorbar(aes(ymin=adj_mean, ymax=adj_mean+tips_sd),width=.4,position="identity")+ geom_text(position = position_stack(vjust = 0.5), aes(label = letter), color = "black",size=4)+ ylab("Number of tips for each branching")+ xlab("Condition")+scale_fill_manual(values=color_branching)+theme(axis.text.x = element_text(angle=90, hjust=1, vjust=1))+labs(fill = "Branching")+ facet_grid(.~genotype)+ theme(axis.text.x = element_text(angle=45, hjust=1, vjust=1))+ theme(legend.position = "bottom", panel.grid = element_blank(), panel.background = element_rect(fill = "white", colour = "black"), panel.border = element_rect(fill = NA, colour = "black"))#+theme(axis.title.x=element_blank(),#axis.ticks.x=element_blank(),axis.text.x=element_blank()) ggsave(here::here(paste0("report/root_architecture/plot/nb_tips_plant_num_branching_all_resumm","19DAT",".svg")),Fig_nb_tips, width = 4.5, height = 6) ```  ```{r, eval=F} Fig1B_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,Angle_ABC2_mean_1), column_value = "Angle_ABC2_mean_1", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, # Ylab_i = expression(paste("Angle ", widehat(ABC2)," (degrees) for order 1")), Ylab_i = paste("Root insertion angle of order 1 (°)"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig1B_stat=Fig1B_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none")+ labs(color="Treatment",fill="Treatment") Fig2B_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,Angle_ABC2_mean_2), column_value = "Angle_ABC2_mean_2", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, # Ylab_i = expression(paste("Angle ", widehat(ABC)," (degrees) for order 2 ")), Ylab_i = paste("Root insertion angle of order 2 (°)"), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig2B_stat=Fig2B_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none")+ labs(color="Treatment",fill="Treatment") # scale_y_continuous(limits = c(50, 100)) # length ################### ---- # for length BC2 Fig3B_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,BC2_mean_1), column_value = "BC2_mean_1", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = expression(paste("Mean of the length of 1st order LR (cm)")), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig3B_stat=Fig3B_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none")+ labs(color="Treatment",fill="Treatment") Fig4B_stat=stat_analyse( data=df_global_archi_select %>% mutate(climat_condition=paste0(water_condition,"_",heat_condition)) %>% mutate(condition=factor(condition,levels=c("Sto_WW_OT","Stoc_WS_OT","Sto_WW_HS","Sto_WS_HS","Wen_WW_OT","Wen_WS_OT","Wen_WW_HS","Wen_WS_HS"))) %>% mutate(climat_condition=factor(climat_condition,levels=c("WW_OT","WS_OT","WW_HS","WS_HS"))) %>% drop_na(climat_condition,BC2_sum_2), column_value = "BC2_sum_2", category_variables = c("climat_condition"), grp_var = "genotype", show_plot = T, outlier_show = F, label_outlier = "plant_num", biologist_stats = T, Ylab_i = expression(paste("Sum of the length of 2nd order LR (cm)")), control_conditions = c("WW_OT"), strip_normale = F, hex_pallet = climate_pallet ) Fig4B_stat=Fig4B_stat[["plot"]] + theme(axis.title.x=element_blank(),axis.text.x=element_blank(),panel.grid.major = element_blank(), panel.grid.minor = element_blank(),legend.position = "none")+ labs(color="Treatment",fill="Treatment") # plot_grid(p_root_projected,Fig1A_stat,p_height_plant,Fig1B_stat, labels = c("A", "B","C","D"), ncol=2, nrow=2) FigB=ggarrange(Fig1B_stat,Fig2B_stat,Fig3B_stat,Fig4B_stat,ncol=4, nrow=1,common.legend = TRUE, legend="right",labels = c("C","D","E","F")) FigBbis=ggarrange(Fig1B_stat,Fig2B_stat,ncol=2, nrow=1,common.legend = TRUE, legend="right",labels = c("C","D")) ggsave(here::here("report/root_architecture/plot/compile_angle_C_F.svg"), FigB,width = 14,height = 3.7) ggsave(here::here("report/root_architecture/plot/compile_angle_C_D.svg"), FigBbis,width = 10,height = 3.7) #svg() # png(here::here(paste0("xp1_analyse/plot/root_architecture/angle_or_root_tips/angle_abc_abd_length_branching_1_2_","19DAT",".png")), width = 30, height = 30, units = 'cm', res = 900) # grid.arrange (FigAngle1) # Make plot # dev.off() # # FigAngle2=ggarrange(img_A,FigAngle1,ncol=2, nrow=1,labels = c("D","")) #compile to del test #FigCompile=ggarrange(FigNbTips,FigB,ncol=1, nrow=2,common.legend = TRUE, legend="bottom") #FigCompile #export to delet # png(here::here(paste0("xp1_analyse/plot/root_architecture/angle_or_root_tips/angle_nb_tips_compile_all_","19DAT",".png")), width = 40, height = 35, units = 'cm', res = 900) # grid.arrange (FigCompile) # Make plot # dev.off() ``` ```{r, eval=F} ################## Compilation of interesting graphs ######################### #FigAngle1=ggarrange(Fig1B_stat,Fig2B_stat,Fig3B_stat,Fig4B_stat,Fig5B_stat,Fig6B_stat, ncol=3, nrow=2,common.legend = TRUE, legend="bottom",labels = c("C","D","E","F","G","H")) #FigAngle1 #export to delet img1 <- readPNG(here::here("report/root_architecture/img/shema_angle2.png")) img_A <- ggplot() + background_image(img1) # png(here::here(paste0("xp1_analyse/plot/root_architecture/angle_or_root_tips/angle_abc_abc'_length_branching_1-2_","19DAT",".png")), width = 30, height = 30, units = 'cm', res = 900) # grid.arrange (FigAngle1) # Make plot # dev.off() tips_img=ggarrange(Fig_nb_tips,img_A,ncol=2,widths = c(0.6, 1), nrow=1,labels = c("A","B")) #compile to del test FigCompile=ggarrange(tips_img,FigB,ncol=1, nrow=2,heights=c(1,0.75), legend="bottom") #FigCompile #export to delet png(here::here(paste0("report/root_architecture/plot/angle_nb_tips_compile_all_","19DAT",".png")), width = 26, height = 22, units = 'cm', res = 900) grid.arrange (FigCompile) # Make plot dev.off() ``` 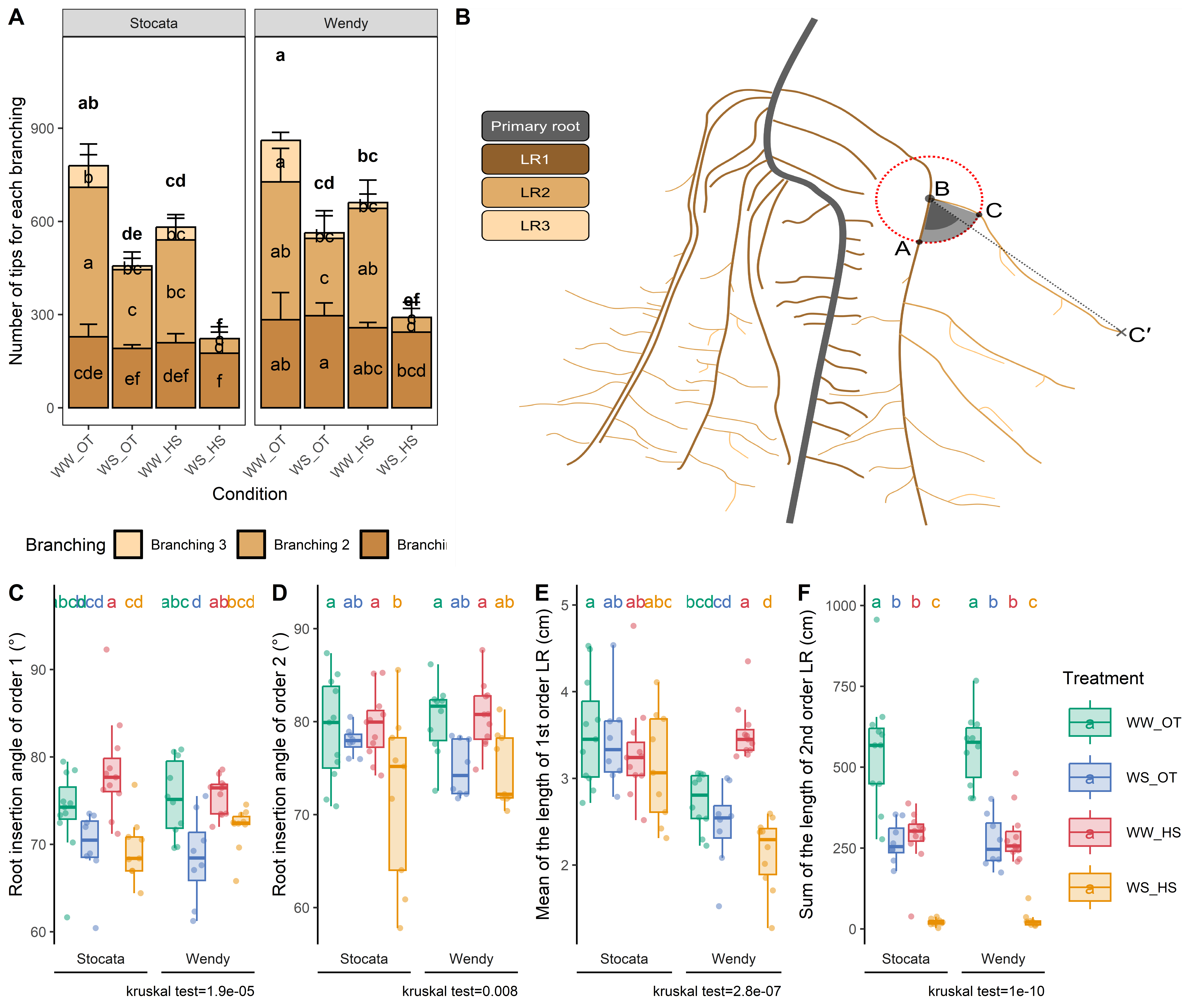 ```{r, eval=F} ################### stats calcule ############ # mean visualisation # df_global_archi_select %>% dplyr::group_by(genotype,climat_condition) %>% drop_na(nb_tips_macro_2) %>% # dplyr::summarise(nb_tips_1=mean(nb_tips_macro_1),nb_tips_2=mean(nb_tips_macro_2),nb_tips_3=mean(nb_tips_macro_3)) %>% as.data.frame() # # student test # variable="nb_tips_macro_3" # # df_global_archi %>% filter(days_after_transplantation==19) %>% # #group_by(genotype) %>% # summarise_each(funs(t.test(.[condition == "Sto_WW_OT"], .[condition == "Sto_WW_HS"])$p.value), vars = variable) # # # means comparison # df_global_archi_select %>% dplyr::group_by(genotype,condition) %>% # drop_na(nb_tips_macro_2) %>% # #filter(genotype=="Stocata") %>% # #filter(climat_condition %in% c("WW_OT","WW_HS")) %>% # dplyr::summarise(nb_tips_2=mean(nb_tips_macro_2)) %>% as.data.frame() %>% mutate(variation_geno=(nb_tips_2[condition=="Stocata_WW_OT"]/nb_tips_2[condition=="Stocata_WW_HS"])) # # # rate of change # df_global_archi_select %>% dplyr::group_by(climat_condition) %>% # drop_na(nb_tips_macro_2) %>% # #filter(genotype=="Stocata") %>% # #filter(climat_condition %in% c("WW_OT","WW_HS")) %>% # dplyr::summarise(nb_tips_2=mean(nb_tips_macro_2)) %>% as.data.frame() %>% mutate(tx_variation_geno=((nb_tips_2[climat_condition=="WS.OT"]-nb_tips_2[climat_condition=="WW.OT"])/nb_tips_2[climat_condition=="WW.OT"])) # # ############## angle stats ############# # df_global_archi_select = df_global_archi %>% filter(days_after_transplantation==19) # #student test # vars_to_test <- df_global_archi_select %>% dplyr::select(starts_with("Angle")) %>% # #dplyr::select(ends_with("2"))%>% # colnames() # # test=df_global_archi_select%>% # summarise_each(funs(t.test(.[condition == "Sto_WW_OT"], .[condition == "Sto_WW_HS"])$p.value), vars = vars_to_test) # # colnames(test)=vars_to_test # # m_angle1=lm(data=df_global_archi_select,Angle_ABC_1~genotype*water_condition*heat_condition) # m_angle2=lm(data=df_global_archi_select,Angle_ABC_2~genotype*water_condition*heat_condition) # summary (m_angle1) # summary (m_angle2) # # df_global_archi_select %>% dplyr::group_by(climat_condition) %>% # drop_na(Angle_ABC2_2) %>% # #filter(genotype=="Stocata") %>% # #filter(climat_condition %in% c("WW_OT","WW_HS")) %>% # dplyr::summarise(Angle_ABC2_1_mean=mean(Angle_ABC2_2)) %>% as.data.frame() %>% mutate(tx_variation_clim=((Angle_ABC2_1_mean[climat_condition=="WS_HS"]-Angle_ABC2_1_mean[climat_condition=="WW_OT"])/Angle_ABC2_1_mean[climat_condition=="WW_OT"])) # # ############## length stats ############# # df_global_archi_select = df_global_archi %>% filter(days_after_transplantation==19) # # #student test # vars_to_test <- df_global_archi_select %>% dplyr::select(starts_with("B")) %>% # #dplyr::select(ends_with("2"))%>% # colnames() # # test=df_global_archi_select%>% # summarise_each(funs(t.test(.[condition == "Wen_WW_OT"], .[condition == "Wen_WS_HS"])$p.value), vars = vars_to_test) # # colnames(test)=vars_to_test # test # # # taux de variation # df_global_archi_select %>% dplyr::group_by(condition) %>% # drop_na(BC2_1) %>% # #filter(genotype=="Stocata") %>% # #filter(climat_condition %in% c("WW_OT","WW_HS")) %>% # dplyr::summarise(BC2_1_mean=mean(BC2_1)) %>% as.data.frame() %>% mutate(tx_variation_geno=((BC2_1_mean[condition=="Wen_WW_HS"]-BC2_1_mean[condition=="Wen_WW_OT"])/BC2_1_mean[condition=="Wen_WW_OT"])) ``` ## Contribution of each variable ```{r, eval=FALSE} df_global_archi_select_v=df_global_archi %>% filter(days_after_transplantation==19) %>% filter(recolte==2) %>% dplyr::select(-c(shooting_date, plant_num ,position ,storage_line, unit, date_recolte,pool_BM, pool_iono, induct_error_evapotranspi,induct_error_root_architecture, note ,plant_letter,recolte)) %>% pivot_longer(-c(plant_num_shooting_date ,genotype,heat_condition,water_condition,condition,days_after_transplantation,climat_condition),names_to = "variable") df_input=df_global_archi_select_v compile_result=as.data.frame(matrix(data=NA,nrow = 0, ncol = 8)) for (i in 1:length(levels(as.factor(df_input$variable)))){ variable_l_i=as.character(levels(as.factor(df_input$variable))[i]) cat( "Variable:" ,variable_l_i, "\n") df_x= df_input %>% filter(variable==variable_l_i) %>% drop_na(value) formula_string <- as.formula(paste("value", "~", paste("genotype","*","water_condition","*","heat_condition", sep = ""))) # Perform ANOVA using the aov function result_variable <- aov(data = df_x, formula = formula_string) anova_result=summary(result_variable)[[1]] %>% mutate(R2 = `Sum Sq` / sum(`Sum Sq`)) row_name<-rownames(anova_result) row_name <- gsub("genotype","Genotype",row_name) row_name <- gsub("water_condition","Water",row_name) row_name <- gsub("heat_condition","Heat",row_name) rownames(anova_result)<-row_name anova_result$variable=as.character(df_x$variable)[1] compile_result=rbind(compile_result,anova_result) } compile_result<-compile_result %>% as.data.frame() %>% rownames_to_column("contribution_variable") %>% mutate(Significance = case_when( `Pr(>F)` <= 0.001 ~ '***', `Pr(>F)` < 0.01 ~ '**', `Pr(>F)` < 0.05 ~ '*', `Pr(>F)` < 0.1 ~ '.', TRUE ~ ' ' )) compile_result=compile_result%>% filter(str_detect(contribution_variable, "Total", negate = TRUE)) %>% filter(str_detect(contribution_variable, "Residual", negate = TRUE)) %>% mutate(contribution_variable = str_replace_all(contribution_variable, " ", "")) %>% mutate_at(vars(contribution_variable), ~str_replace_all(., "\\$", ""))%>% mutate_at(vars(contribution_variable), ~str_replace_all(., "[0-9]", "")) %>% mutate(contribution_variable = as.character(contribution_variable)) %>% mutate(fontcolor = ifelse(contribution_variable %in% c("Water", "Genotype:Water:Heat"), "#ffffff", "#000000")) %>% mutate(contribution_variable=fct_relevel(contribution_variable,c("Genotype","Water","Heat", "Genotype:Water","Genotype:Heat","Water:Heat","Genotype:Water:Heat"))) %>% mutate(text_output=paste0(round(R2*100,0), "% ", Significance)) compile_result =compile_result %>% drop_na() plot_contrib<-ggplot(compile_result, aes(x = variable, y = R2, fill = contribution_variable)) + geom_bar(stat = "identity") + geom_text(aes(label = ifelse(R2 > 0.02, text_output, "")), color = compile_result$fontcolor, position = position_stack(vjust = 0.5), size = 2.5)+ scale_fill_manual(values = c("#ffd166", # geno "#118ab2", # water "#ef476f", # heat "#06d6a0", # watergeno "#f78c6b", # genoheat "#FFB6C1", # waterheat "#333333"), name = "title of legend (contribution") +#all scale_y_continuous(labels = scales::percent_format())+ scale_color_identity()+ labs(x = "Compartment", y = "The relative contribution for the different variables (%)", ) + theme_minimal()+ theme(panel.grid = element_blank(), legend.title = element_blank(), #axis.text.x = element_text(angle = 45,hjust = 1, vjust = 1), axis.title.x = element_blank(), legend.position = "bottom" ) + coord_flip() ggsave(here::here("report/root_architecture/plot/contribution.svg"), plot_contrib, height = 13,width = 12) ```  


